Fine-Tuning Small Domain Models: When SLMs Outperform LLMs
Introduction
Massive general-purpose language models like GPT-4 have dominated the AI landscape, impressing with their broad knowledge and fluency. But bigger isn't always better for every task. In highly specialized domains, a smaller model fine-tuned on domain-specific data can outshine a giant pre-trained model that lacks specialization. arXiv:2305.07804 is a good starting point for this hypothesis. The authors show that a carefully fine-tuned Small language Model (<1.6B parameters) can outperform bigger models like GPT-4 on narrowly defined tasks. This article explores that phenomenon through our own experiment: we fine-tuned a LLaMA 8B model on a biomedical Q&A dataset and found that it significantly outperformed GPT-4 on those domain-specific questions, despite GPT-4's larger size.
Why does this happen, and what does it mean for those of us without unlimited budgets? In this post, we'll dive into the experiment, discuss results, and highlight how fine-tuning a smaller model for <$5 can sometimes beat using a large general model that costs much more to operate. Our goal is both technical and educational: to broaden your perspective on AI deployment options. Large Language Models (LLMs) aren't the only option – with the right tools, Small Language Models (SLMs) tailored to your data can be cheaper, more private, and even more accurate for your specific use case.
Background: Large vs. Small Models in Specialized Tasks
General LLMs like GPT-4 are trained on internet-scale data, giving them a very broad (but not always deep) knowledge of many domains. They excel at a wide range of tasks out-of-the-box. However, when it comes to deep, precise understanding within a niche domain, a model that has been specifically fine-tuned on that domain's data often has an edge in both accuracy and efficiency. Fine-tuning essentially teaches a smaller model the exact patterns and knowledge needed for a particular task, which a general model migt only know superficially.
There are some clear trade-offs between using a large generic model versus a small fine-tuned model:
- Accuracy in Domain Tasks: A well-tuned domain-specific model tends to outperform a general model like GPT-4 in that domain. The specialization allows it to answer with confidence where GPT-4 might hallucinate.
- Cost and Efficiency: Using GPT-4 via API has a pay-as-you-go cost based on tokens. For heavy or complex queries, this can become quite expensive. In contrast, once you fine-tune your Small Language Model, you can deploy it locally which can cut costs exponentially.
- Data Privacy: With a hosted API like OpenAI's, your prompts are sent to a third-party server. A custom model that you fine-tune and run locally or on a private cloud gives you full control over your data, a key consideration for healthcare, finance, and other sensitive domains.
- Flexibility and Control: Fine-tuning allows you to imbue the model with a specific tone**,** style, or constraint relevant to your task. A generic model might not follow your preferred format whereas a fine-tuned model can be trained to do exactly that.
Of course, training a domain model has upfront costs (preparing data, running the fine-tuning process) and requires expertise. But this is exactly where Radal comes in! We're building a no-code, drag-and-drop platform to make fine-tuning easy and cheap. Our platform lets you visually create a training pipeline, upload your dataset, set some parameters, and fine-tune a model in a few clicks. The goal is to allow everyone, including non-technical users to reap the benefits of custom AI models without deep ML knowledge. And as we'll show, even a <$5 fine-tune can yield significant gains for specialized tasks.
The PubMedQA Challenge Dataset
For our experiment, we chose the PubMedQA dataset – a benchmark for biomedical question answering. PubMedQA consists of research questions (often derived from article titles) that must be answered with "yes", "no", or "maybe". Crucially, each question is tied to an abstract from the biomedical literature, which contains the evidence needed to determine the answer (yes/no/maybe) based on the study's conclusions. What makes PubMedQA particularly challenging is that it requires reasoning over complex research texts – often involving understanding study design, outcomes, and even quantitative results – to infer the correct answer. It's not just simple fact lookup or common-sense reasoning; the model has to act like a domain expert analyzing research data.
To give a sense of difficulty, more than 20% of questions require subtle reasoning that stumps typical models. This "yes/no/maybe" format is also tricky because "maybe" is a legitimate answer. A model must not only know facts but also judge the certainty of evidence. All these factors make PubMedQA an excellent stress-test for domain understanding. It's the kind of task where an untrained general model might struggle, but a model specialized on biomedical text could excel.
For our purposes, we treated PubMedQA as a closed-book QA task: we wanted to see if models could answer the questions from their internal knowledge after training, without being given the relevant article text at test time. This is even harder than the standard task, where the model would typically get to read the abstract before answering. We chose this setup to simulate a real-world scenario like asking a medical AI a question and expecting it to know the answer or at least not hallucinate. It also emphasizes the knowledge vs. reasoning aspect – can the model recall or deduce the answer just from the question context?
Experiment Methodology
We fine-tuned a LLaMA 8B parameter language model on the PubMedQA dataset's Q&A pairs. The fine-tuning process involved training the model on many question, answer examples with their ground truth labels. Essentially, we taught the model how biomedical questions are answered based on research findings. The fine-tuning was done using our no-code pipeline, and it was remarkably quick and cheap – the entire fine-tune job cost under $5 in compute time. This low cost is due to the combination of a relatively small model (8 billion parameters is tiny compared to GPT-4's estimated 170B+) and a manageable dataset size (PubMedQA has 1,000 expert-labeled examples, plus we could leverage some of the 61k unlabeled or 211k synthetic QAs if needed ).
For testing, we took 100 randomly selected questions from PubMedQA test split. We then queried three models with each question, using an identical prompt format for fairness:
- Our fine-tuned LLaMA 8B model (after training on PubMedQA).
- GPT-4 (OpenAI API), using the latest GPT-4 model.
- LLaMA 8B base model (the original pre-trained model without any fine-tuning).
We used the same 100 questions for each model. In all cases, we did not provide the models with the supporting abstracts or any additional context. This meant GPT-4 had to rely on whatever it knows about biomedical research, and similarly for the base LLaMA. The fine-tuned model, on the other hand, might have internalized some patterns or facts from PubMedQA during training that could help it answer.
We recorded each model's answer and then compared it to the ground truth labels from the dataset. Accuracy was calculated as the percentage of questions for which the model's answer exactly matched the ground truth.
We also logged the reasoning each model gave which was not used for scoring, but it provided insight into why the model gave a certain answer. In many cases it highlighted differences: GPT-4 would often produce a very general explanation, whereas the fine-tuned model sometimes gave concise domain-specific justifications.
Inference for LLaMA models was done via the LM Studio local endpoint, while GPT-4 was accessed through the OpenAI API. Despite the vastly different infrastructure, we attempted to keep conditions as similar as possible.
Results: Fine-Tuned Model vs. GPT-4 vs. Base Model
The fine-tuned LLaMA 8B model delivered the highest accuracy on the 100 biomedical questions, outperforming GPT-4 by a notable margin. The base LLaMA trailed slightly behind GPT-4. Here's a summary of the accuracy we observed:
- LLaMA 8B fine-tuned on PubMedQA: 26.8% accuracy (correct on 26–27 out of 100 questions).
- GPT-4 (OpenAI, general model): 16.0% accuracy (correct on 16 out of 100).
- LLaMA 8B base (no domain training): 14.0% accuracy (correct on 14 out of 100).
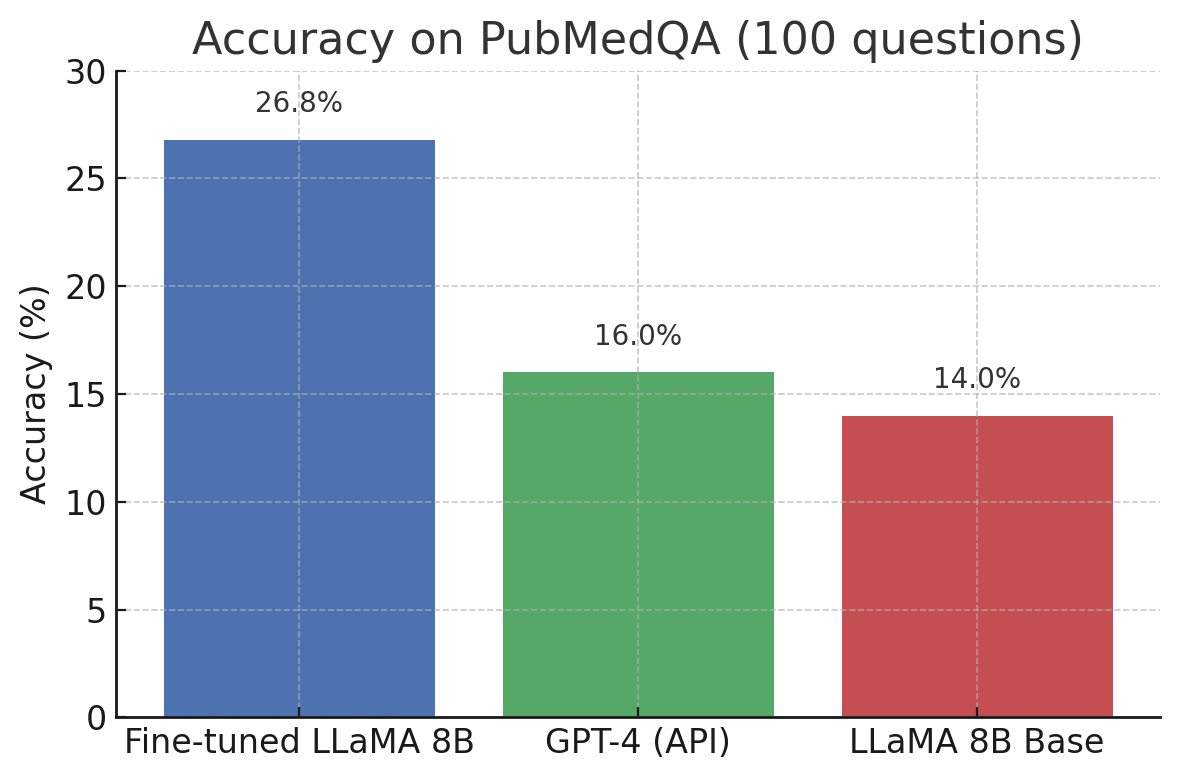
Accuracy of each model on 100 PubMedQA questions. The fine-tuned 8B model answered ~27% correctly, significantly surpassing GPT-4's 16% on this domain-specific task. The base 8B model (no fine-tuning) achieved 14%. Although the fine-tuned model's absolute accuracy is modest, it clearly outperforms the much larger GPT-4 when answering specialized biomedical questions.
These results support our hypothesis: a small model trained on domain-specific data can beat a state-of-the-art large model on those specific tasks. The fine-tuned model answered about 10 more questions correctly (out of 100) than GPT-4 did. Interestingly, the base LLaMA model, which had never seen the PubMedQA data, got nearly the same number right as GPT-4 (14 vs 16). GPT-4's advantage over base LLaMA in this setting was minimal – suggesting that without domain adaptation, even a powerful general model struggles with these kinds of questions.
Why did GPT-4 perform so poorly here? One stark observation was GPT-4's tendency to answer "maybe" to almost everything. In fact, GPT-4 responded "maybe" for approximately 80% of the questions in our sample. This implies that when uncertain, GPT-4 defaulted to the safest-sounding option: maybe. The problem is that "maybe" was only the correct answer for 4 out of the 100 questions (in our sample, the ground truth distribution was heavily skewed: 68 "yes", 28 "no", and only 4 "maybe"). Thus, GPT-4's strategy of frequently saying "maybe" completely bombed its accuracy. It suggests that GPT-4 did not know the answers to most of these specific medical research questions, and because it "knows it doesn't know," it hedged. This is a form of hallucination avoidance – rather than hallucinate a yes or no, it often gave a non-committal answer.
The base LLaMA 8B model likewise showed uncertainty, answering "maybe" 85% of the time in our test. Clearly, a base 8B model doesn't have the knowledge to address these questions confidently either. Its 14% accuracy likely came from lucky guesses or any generic training knowledge it had.
In contrast, our fine-tuned LLaMA 8B had a very different answer pattern. It answered "yes" or "no" far more often, and reserved "maybe" for fewer cases. This indicates the fine-tuning taught it a certain bias towards giving a definitive answer. In fact, it may have overcorrected – our fine-tuned model said "no" much more than "yes", whereas the actual answers were often "yes". This mismatch likely capped its accuracy. With further tuning or calibration, we could perhaps improve its yes/no balance and get higher accuracy. Nonetheless, the fine-tuned model demonstrated knowledge and patterns from the domain that GPT-4 lacked. For example, on a question about whether health information exchange reduces redundant medical imaging, the fine-tuned model correctly answered "yes" (and cited reasoning about studies showing fewer duplicate tests), whereas GPT-4 said "yes" as well in that case – but on many other questions where the answer was "yes", GPT-4 defaulted to "maybe" and got it wrong, while the fine-tuned model often knew to say "yes" or "no" as appropriate.
It's worth noting that the absolute accuracy numbers are low in an absolute sense. 27% correct is a long way from the 68% that specialized research models have achieved on PubMedQA . This gap is because our fine-tuned model is still relatively small and because we gave it a very minimal fine-tuning regimen. Moreover, we didn't provide the actual abstracts at test time – we essentially asked the model to recall or reason out answers without the supporting text, which is extremely challenging. So, don't interpret 27% vs 16% as an indictment of GPT-4's overall capabilities – if we had given GPT-4 the relevant article text to read for each question (as the task is formally defined), it would likely score much higher (perhaps closer to the 70-75% range. Our focus was specifically on knowledge and specialization: with no additional context, the small fine-tuned model clearly had more built-in knowledge of these specific Q&As than GPT-4 did.
Discussion: Why Small Domain Models Can Win (and When to Use Them)
Our experiment reinforces a key point in the emerging LLM landscape: for narrow tasks, a smaller model that's trained on domain-specific data can be more effective than a large general model . This is analogous to human experts – a general practitioner doctor knows a little about many conditions, but a cardiologist will outperform the general practitioner on a specific heart-related question.
There are several reasons why the fine-tuned small model outperformed GPT-4 here:
- Domain-Specific Knowledge: Through fine-tuning, the LLaMA 8B effectively absorbed details from the PubMedQA dataset – including how certain conditions relate, what outcomes studies found, etc. GPT-4 was never explicitly trained on that dataset (to our knowledge) and so it had to fall back on general medical knowledge. If PubMedQA's questions involve nuanced or less-common research results, GPT-4 wouldn't reliably know those. The small model, however, memorized or generalized from those exact Q&As during fine-tuning.
- Optimization for the Task: We explicitly trained the 8B model to output only "yes", "no", or "maybe" with the reasoning. GPT-4, on the other hand, is a general conversational model. Even with a system instruction to answer in one word, it sometimes would diverge or qualify its answer. The fine-tuned model had no qualms about giving a one-word answer because that's how it was trained. This kind of format specialization is a small but important advantage in getting accurate classification-style outputs.
- Avoiding Over-Cautiousness: As noted, GPT-4's helpfulness and caution may have led it to say "maybe" too often. The fine-tuned model learned from the training data frequency that "maybe" is actually a less common correct answer, so it was more willing to choose yes or no. In essence, the fine-tuning corrected the prior that "maybe" is rarely the right choice unless there is evidence of an inconclusive result. GPT-4 didn't have that calibrated, so it chose "maybe" whenever uncertain – which in this case was frequently wrong.
From a cost-benefit perspective, the implications are exciting: our fine-tuning run cost on the order of $5 (for 1,000 examples, a few epochs on 8B parameters using a cloud GPU instance). Running the fine-tuned model for inference is also cheap – it can even be done on a decent consumer GPU or CPU at a few cents worth of electricity per 100 questions. By contrast, querying GPT-4 for 100 questions would cost quite a bit. If we scaled this up to thousands of questions or a deployed application answering user queries, GPT-4's costs would accumulate quickly, whereas a self-hosted 8B model remains fixed-cost and could handle many queries per second without breaking the bank. In scenarios where you need to answer a high volume of domain-specific queries, fine-tuning a model and self-hosting it can yield huge cost savings.
There are caveats, of course. Fine-tuned small models are narrow experts: our PubMedQA-trained LLaMA 8B is great at PubMedQA-style questions, but ask it something outside that distribution and it might fail spectacularly or spout nonsense. GPT-4, however, could gracefully handle a much wider array of questions. Thus, for broad applications or when you can't clearly define the domain, a general model might still be the best choice. There's also the maintenance aspect: if new research comes out, our fine-tuned model wouldn't know about it unless we update the training data and retrain. GPT-4 might catch some new information if it's updated or via retrieval plugins.
However, these challenges are surmountable with a good MLOps pipeline and are often acceptable trade-offs given the benefits. For many real-world use cases – say a medical assistant tool for clinicians, a legal document analyzer, a customer support FAQ bot – the questions you need to answer are within a particular domain and format. You can define that scope, collect data for it, and fine-tune a model. Our experiment shows that even a relatively small model can then achieve domain-specific competence that might rival or exceed a large model.
Conclusion
Our first experiment in this series demonstrates a clear proof of concept: with minimal cost and effort, a fine-tuned 8B parameter model beat one of the world's most advanced large models (GPT-4) on a task of answering specialized biomedical questions. The smaller model's domain-focused training gave it an edge in accuracy and the ability to provide more decisive answers, whereas the larger model often wavered despite its greater general knowledge.
For practitioners and businesses, the takeaway is don't assume you must default to a giant LLM for every problem. If you have a well-defined task and a good dataset, consider fine-tuning a smaller open-source model. It could not only boost performance for that task but also save costs, improve privacy, and allow custom behavior that a generic model won't offer.
Broader Perspective: Large, general AI models are amazing generalists, but specialists can beat generalists in their own arena. Just as you wouldn't use a Swiss Army knife for a job that requires a scalpel, it's time to recognize that a smaller, well-honed model might serve your needs more effectively than an expensive universal model. And now, even those without technical backgrounds can wield that scalpel, using intuitive tools to train and deploy custom models. We hope this exploration inspires you to experiment with SLMs for your projects.